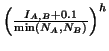 where
where There is only one method in this class which must be provided/implemented by the user, and it is evaluateOutput() (marked with a *).
=> OBJECT, ExperimentId => STRING, DBFileStem => STRING, ATTRIBUTE-HASH (optional)
As with the Algorithm constructor, you probably don't need to call this unless you are developing specialist applications. The obligatory arguments are: Population, ExperimentId and DBFileStem. For some applications it is OK to give a dummy value for Population.
As with Algorithm, this is a cascaded method where the attributes are set. The defaults are defined in GeneticProgram::_init() and you should customise the object by editing attribute/value pairs in Individual::_init() because these override all defaults.
This doesn't do much in itself, but is a very important wrapper method - to be called whenever evolved Individual-specific methods need to be redefined.
This is where the Perl code is actually evaluated, in order to redefine certain Individual-specific subroutines, commonly evaluateOutput() and evolvedInit(). The code is obtained using getCode() and simply passed to Perl's eval() function. Before that, a signal handler is set to filter out ``Subroutine redefined'' warnings.
If an error occurs during the eval(), this usually means that there is a syntax error in the evolved code (usually just subroutine definitions, nothing is actually executed), caused by a mistake in the Grammar. Warnings are printed to STDERR and the Individual is randomly reinitialised with a new genome before attempting evalEvolvedSubs() again (with a 15 second sleep in between). The user should fix these syntax errors to make sure valid Perl is generated in all circumstances.
The supplied method does nothing, but you can provide an evolved version to set some of the Individual's attributes. The sine curve demo in Section 9.2 shows how to have evolvable mutation and crossover rates. Note that reInitialise() and hence evolvedInit() is called inside mutate() and crossover() in anticipation that you might use evolvable mutation and crossover parameters.
This is where you should provide the code which works on inputdata and produces outputdata. Normally you would setup up the Grammar so that evaluateOutput() is defined in the code-tree and is therefore evolved, but you could define evaluateOutput() as a non-evolvable function and have it call other evolved functions.
As discussed in Section 3, the scalar variables inputdata and outputdata are usually references to arrays or hashes containing multiple data points. It is entirely up to you how you represent the data, since you are in control of reading it in (with Algorithm::loadSet()), generating the output (in evaluateOutput()), assessing the fitness (in Algorithm::fitnessFunction()) and writing it out (in Algorithm::saveOutput()).
Here you can specify which extra information about the best-of-tournament Individual will be saved in `results/tournament.log'. The default function just returns DBFileStem, but you could have it return evolved parameters, for example. The sine demo described in Section 9.2 uses this technique.
This method finds crossover points between the self object and the mate object. If FixedXovers is set and FixedXoverProb is satisfied, then FixedXovers pairs of crossover points will be selected. Otherwise the default and recommended behaviour is to use NodeXoverProb to calculated the number of crossovers to attempt, as described for in the following pseudocode:
crossovers_to_do = 0
for i = 1 to number_of_nodes_in_tree {
if (rand(1) < NodeXoverProb) {
crossovers_to_do++
}
}
Crossover point selection is a little bit complex, so only an outline is given here. The general approach is to sample pairs of points until they satisfy certain criteria. To prevent infinite sampling maximum number of samples are attempted (equivalent to sampling each node 100 times). One node each from the self and mate genome trees are selected randomly (with depth bias XoverDepthBias); we will call them mynode and matenode in the following discussion. They are accepted if all the following conditions are met (in the order given):
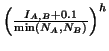 where
where After the crossover points have been selected, they are passed to _start_crossover(). If no crossover points were found (or if none were requested) then the two parents are crossed over at the root node, which is equivalent to asexual reproduction. You can also set AsexualOnly to force asexual reproduction.
With probability XoverLogProb, some information about crossover points is written to XoverLogFile.
A wrapper around the recursive _crossover() method, which first does a tieGenome() on the recipient and wipes its genome.
Recursive subroutine which builds up a new genome tree for recipient by tracing through the self genomes and switching over to the mate genome when a crossover_pair is met. To avoid name clashes mate nodes are copied with names appended with an ``x'' (see also _xcopy_subtree()) and these are tidied up by _fix_nodes() on returning to _start_crossover().
This method does one round of mutation on an Individual. First the number of mutations to be attempted is calculated according to the following pseudocode:
mutations_to_do = 0
for i = 1 to number_of_nodes_in_tree {
if (rand(1) < NodeMutationProb) {
mutations_to_do++
}
}
However if FixedMutations is non-zero and FixedMutationProb is satisfied, then FixedMutations are attempted.
Each mutation is randomly chosen to be either point mutation (via method point_mutate_shallow()) or a macro mutation (via method macro_mutate()), according to the attribute PointMutationFrac.
If NoNeutralMutations is non-zero, then a check is made that the mutation made a visible (not necessarily functional) change to the Perl code. This is done by expanding the genome-tree before each mutation, and after each mutation and comparing the two strings. If the strings are identical then another mutation is attempted (up to a safety limit of 5000 tries). This feature is turned off by default.
Note that we say that mutations are attempted rather than performed. That is because some of the mutation operators fail to find suitable nodes to act on, and as a result do nothing.
After the mutations are are done, some logging is done with probability MutationLogProb into a file MutationLogFile, with some simple information about the nodes that were mutated (if they still exist - when more than one mutation is attempted, the subsequent mutations can delete the nodes affected by previous mutations).
If more than one mutation was attempted, then the cached fitness of the Individual is reset with a call to initFitness().
Simple wrapper to call point_mutate() with depth bias (PointMutationDepthBias) more in favour of leaves.
Simple wrapper to call point_mutate() with depth bias (MacroMutationDepthBias) more in favour of internal nodes.
Attempts to make a point mutation, that is a change in the genome-tree which does not affect the branching pattern and involves just one node. The first thing it does is select a random node with depth bias depthbias.
=> NUMBER, start_node => STRING (optional), not_this_node => STRING (optional), not_this_subtree => STRING (optional), node_type => STRING
Picks one node from the genome-tree according to the following pseudocode:
N = number_of_nodes_below(root_of_tree)
do {
node = pick_a_random_node_from_tree
S = number_of_nodes_below(node)
} until rand(depth_bias * N) <= S
In this way, it is possible to vary the amount of bias towards internal nodes. With a depth_bias of zero, all nodes are selected with equal probability, but with a depth_bias of 1 only the root node will be selected with absolute certainty. Of course, it is not very time-efficient to sample nodes in this way.
Other optional parameters allow a specific node type to be selected, or nodes within or outside a certain subtree. If a node cannot be found after 5000 tries, the routine gives up and returns the empty string.
Picks at random one of the available macro mutation operator names
from the array MacroMutationTypes and invokes the method of that name (in Perl you can call a method using a string variable like this: $method = 'swap_subtrees'; $individual->$method()).
Picks a random node with MacroMutationDepthBias and deletes the subtree and ``grows'' a new subtree in its place.
Inserts a single new function node internally in a tree.
First we pick a random node with MacroMutationDepthBias and copy its contents to a brand new node. The original node is then replaced with a random function with at least one branch point of the same type as the original node (if none exists, then we give up). We link up one of these branch points back to the new node, and any remaining branch points have new subtrees grown onto them. Figure 1 illustrates this complicated procedure.
![\includegraphics[width=0.66\textwidth]{figures/insert_internal}](img12.png)
|
Selects a node with MacroMutationDepthBias, and then, if possible, a second node of the same type within the subtree of the first. The two nodes are reconnected and the intervening nodes are deleted (actually it is implemented in another way, but the result is the same).
Selects a node with MacroMutationDepthBias, and then, if possible, a second node of the same type outside the subtree of the first. Deletes any sub-nodes belonging to the second node and copy the first node's subtree to the second node.
Like copy_subtree, except that no nodes are deleted, the subtrees exchange positions.
An experimental, not fully tested method which takes a subtree (which represents not more than fraction EncapsulateFracMax of the whole tree) and replaces it with a single terminal node containing the expanded code of the former subtree. The code is passed through simplify() to remove any redundancy (if you implement the method). The new node is tagged with a special prefix string `;;NUM;;' where `NUM' is the size of the subtree before encapsulation. This number is used in _tree_type_size() (which is sometimes called by getSize()) during tree size calculation to make it fair when parsimony is used. As I said, this needs a thorough investigation because things have changed since I last used encapsulation, and it is a pretty complicated operator with many side effects.
The default method does nothing, but you can define a method to do search-and-replace-with-eval simplification, for example:
sub simplify {
my ($self, $code) = @_;
my $z = 0;
while ($code =~ s/(\d+ \+ \d+)/eval $1/e) {
last if ($z++ > 1000); # safety measure
}
return $code;
}
will simplify `1 + 2 + 3 + 4' to `10'. This can be very powerful for creating human-readable output.
A helper method which takes a node and generates a copy of its subtree with new nodekeys which are the same as the original tree plus a trailing `x' (so `nodeNUM23' is copied to `nodeNUM23x'). This is a simple way to avoid node key clashes, as long as you call _fix_nodes() afterwards...
Recursive method to fix the nodekeys which end in `x' and replace them with valid nodekeys which don't end in `x'. It searches incrementally for new unused nodekeys from zero (in other words, `nodeNUM14x' is replaced by `nodeNUM0' and if that already exists, then `nodeNUM1', `nodeNUM2' ...until a free key is found).
Returns the nodekeys of all subnodes of the given nodekey.
Recursive method which ``on the way out'' deletes all the nodes in the given subtree.
This method descends a subtree (usually `root') and fills a number of hashes (actually the hash references given as arguments: sizes, types) with the size of the subtree below each node, and the nodetype of each node (the hash keys are nodekeys).
If the ignore hash reference is defined, then this routine will not descend subtrees of any nodetype which exists as a key in the ignore hash.
The methods crossover() and mutate() efficiently make use of the cached size information when sampling nodes and repeated tree recursion is not needed.
Prints a text dump of the nodes below nodekey to STDERR for debugging purposes.
=> STRING, Tournament => NUMBER
Prints the Perl code and some other relevant information to Filename for the user.
=> STRING, StartNode => STRING, HighLight => HASH-REF
Prints the genome tree to Filename in a format readable by the third party program daVinci. The optional hash reference HighLight can be used to give certain background colours to certain nodes (key = node-id, value = hex colour).
=> STRING
Copies the genome hash to a new DB file with FileStem.
=> STRING
Loads up a genome hash from a DB file with FileStem. Erases the current genome, but consider making a call to reInitialise() to update evolved parameters afterwards.
By default, an Individual is not ``connected'' to the DBM file
which stores the genome. If it was, then a typical run would
have thousands of open filehandles and would probably crash.
This method uses Perl's tie() to tie the hash stored
in $self->{genome} to the file(s) identified by DBFileStem. This is
done in read/write/create mode. See the Perl documentation
for tie() for more details.
The return value is a reference to the genome hash, or you
can use $self->{genome} if you prefer.
Make sure you always have a corresponding call to untieGenome() to avoid filehandle overload. Symmetrical nesting of ties and unties is allowed - if the genome is already tied a counter is incremented in tieGenome() which is decremented by untieGenome() - the actual untie is done only at ``depth zero''.
Note that you don't want to have to many tie/untie tie/untie tie/untie sequences (big performance hit), it's better to wrap a tie...untie around them all. This is why there are tieGenome() calls in TournamentGP::crossoverFamily(). If you can't figure out where/when/why various tie/untie calls are not made, you can uncomment the debugging line and watch STDERR.
If you want to completely wipe the genome, see the source code for _start_crossover().
See tieGenome().
The DBM files seem to grow with time, so this method will copy the genome hash into memory, and then delete the DBM files, and rewrite a new one. Obviously this is a bit risky since there is no backup made on disk.
Currently only called in _start_crossover() on the recipient/child after wiping the genome.
Does not affect the tie level counter (see tieGenome()).
This is a simple wrapper around _init_tree() which does tieGenome() first and untieGenome() afterwards.
Initialises the genome hash and starts building a new tree of max depth TreeDepthMax. Repeats this until the number of nodes is greater than MinTreeNodes.
=> INTEGER,
type => STRING,
TreeDepthMax => INTEGER
Builds a tree or subtree by randomly selecting functions and terminals. TreeDepthMax is passed as an argument (the object attribute of the same name is not used) because you might want to use different limits for the whole tree vs. subtrees. The depth argument is incremented on each recursive call. Usually a function is chosen during tree building but if the depth is greater than TreeDepthMax then a terminal is always added. Alternatively you may use TreeDepthMin and TerminateTreeProb to force termination with some probability below a vertain depth.
This method expands the tree below nodekey into a single string which I presumably Perl code (or at least something that will eval()). It is done with an iterated search-and-replace. A counter keeps track of how many nodes have been expanded and if this is greater than MaxTreeNodes then no more tree expansion is done and random terminals are used to ``finish off'' the expansion to generate valid, but presumably unfit, Perl code.
You should probably use the public method getCode(), which wraps this.
Recursive tree-descending method which goes down two subtrees and counts the number of nodes identical between them at equivalent topological positions. It stops descending whenever nodes are unequal, therefore it is quite crude and cannot handle insertions or deletions.
Deletes the two cached copies of the fitness (one in the genome, one in memory).
Deletes all contents of the $self->{memory} hash, which contains the cached fitness value, but you
may also store other things here, using the memory()
method.
A get and set method for the $self->{memory} hash. With a single argument, it returns the value
stored with that key. With an even number of arguments
(in other words, a hash) it sets the key/value pairs.
If you prefer, you can use getMemory() and setMemory() which are simply wrappers to this method.
This is the main method for getting the Perl code from the genome.
This is the method for calculating the size of the genome. If the attribute GetSizeIgnoreNTypes is used then _tree_type_size() is used to calculate the number of nodes by descent (ignoring nodes which lie below the nodetypes defined by GetSizeIgnoreNTypes). Otherwise, the number of keys in the genome hash is returned (which is much quicker of course).
Without an argument routine returns the Individual's Fitness, first from the memory hash (see memory()) or if that is undefined, then a tieGenome() is called and the fitness value is retrieved from disk, assigned into the memory cache, and returned. Note that ``undefined'' value for Fitness usually signifies that that the fitness has not yet been evaluated, but sometimes just means that the fitness has not been retrieved from disk.
With an argument, a new value for Fitness is stored in memory and on disk.
Without an argument, it returns the ``age'' of an Individual, which usually means the number of tournaments participated. It is stored only in the memory cache (see memory(), and not on disk.
This routine first calls _random_existing_terminal() and returns its result if true, otherwise it returns one random element from the array stored in the hash pointed to by Terminals using the hash-key nodetype.
If you have numeric mutation switched on, perhaps you want to use existing refined numeric terminals during the construction of new subtrees. If so, you can set UseExistingTerminalsFrac to a fraction/probability, then this method will sometimes return a randomly chosen existing terminal of type nodetype (if one exists) from the genome tree of this organism. For the random selection, each unique terminal is counted only once (to prevent saturation of that terminal).
If UseEncapsTerminalsFrac is non-zero and if the optional argument encapsulated is true, then with this probability, the method will return only terminals of the requested type which were generated by encapsulation (these are tagged with ``;;NUM;;'', see encapsulate_subtree). If none are found, an empty string is returned.
If no set of functions of type nodetype is defined in the Functions, then this function passes through to _random_terminal(). Otherwise, with probability UseEncapsTerminalsFrac a random encapsulated terminal is attempted to be chosen through a call to _random_existing_terminal().
But the default behaviour is, of course, to return a random function of type nodetype from the Functions hash.